News & Events
Health Data Preparation for Artificial Intelligence
22 - FebruaryThe generalizability and reusability of an AI model and digital intervention or decision support service built on that model are some of the most important challenges in scaling up AI solutions in different healthcare settings. Many important factors make this a difficult problem and interoperability is a major one.
An AI solution is generally developed on a specific dataset consisting of several features extracted from a set of data sources within a clinical setting. This dataset should be well defined syntactically and semantically if the AI solution will be validated in other cohorts (reproducibility) and will be used in real life in different care settings. But this is not the case for current research and clinical application practices in the field. This is also the focal point for the FAIR data initiative and principles. Generally, the data preparation process is performed by data scientists and engineers in an offline way for each project which is a cumbersome process and only used in research phases (e.g., preparation of training data, cross-validation data, etc.). When it comes to deploying the resulting AI models and services for daily clinical practice, integration becomes a bottleneck for reusability and exploitation of developed AI models as they are generally tightly coupled with a specific database, file, object store or data stream.
This coupling is also very difficult to maintain as any change in data infrastructure will break the dependent AI solution. Furthermore, different implementations of data retrieval for the training phase and data retrieval for the online serving (prediction) phase leads to inconsistencies in data and result in poor accuracy and reliability. This is one of the reasons why there is not much AI algorithm/model serving at the frontlines of clinical practice and only big companies are dominant in the market.
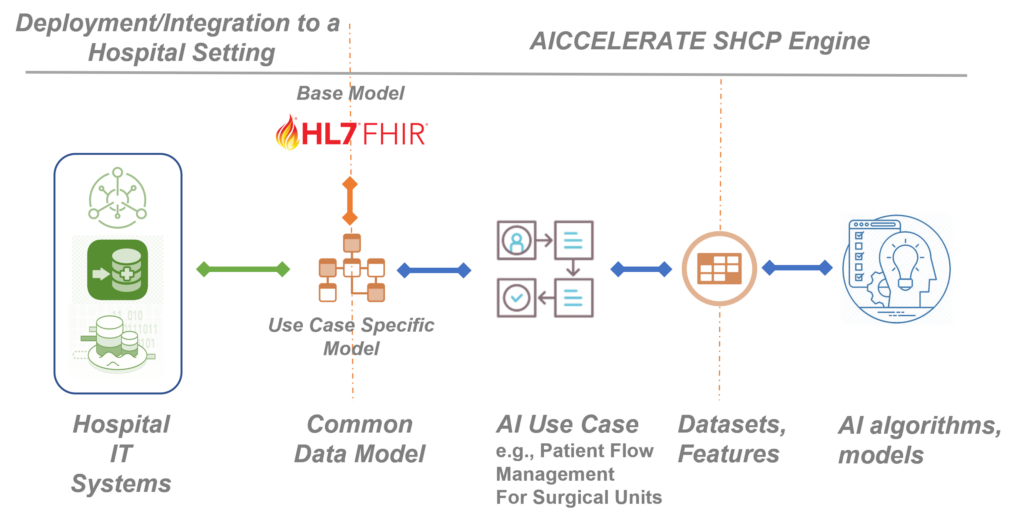
Building on the previous H2020 project results, in the AICCELERATE project, we are developing, validating, and improving a methodology and toolset based on international health data standards, e.g., HL7 FHIR to overcome the interoperability problem by also considering the FAIR data principles. As a first step, rather than starting directly from defining input parameters for an AI model/service, we define a common health data model and data interface with rich machine-processable metadata for the AI use case in hand from the clinical perspective. In other words, we are trying to model the raw data that is produced within the related workflow (e.g., data representing a prescription given to the patient residing in EHR) as this is the integration you need when you want to use an AI model as a decision support service in clinical practice.
HL7 FHIR standard is used as the base data model and its profiling framework is used to define the customizations and extensions required by the given use case. The FHIR standard defines metadata resources for defining data models (data elements restrictions on it including clinical terminology bindings) and data access API. For each use case, these resources are provided to clearly define the data model and the API which in fact will be the single data access interface (plug-and-play port) for a third-party organisation that wants to reuse or exploit the resulting AI solution. This decouples the AI solution from the data infrastructure of the organisation which might be proprietary.
Using an Artificial Intelligence model as a decision support tool in clinical practice requires end-to-end integration starting from the raw clinical data.
If an organisation already opens their health data (e.g., EHR, PHR, a data lake) through an FHIR API, the integration can be easily achieved with some small tunings for conformance with the defined common data model defined as FHIR Profiles. However, this is still challenging for especially small and medium-size care settings. Therefore, we provide a toolset, a Data Ingestion Suite, that enables users to graphically define mappings (ETL definitions) from any source data format to the FHIR based common data model and configure the ETL engine given as a part of the suite for batch or stream processing to deal with the integration. An open-source HL7 FHIR compliant secure health data repository, onFHIR.io, is also provided as a part of the suite which acts as a gateway and provides the defined API with the stored health data transformed via the ETL engine.
Another decoupling is needed between the AI solution and data access layer as there are generally more than one subproject or prediction problem which may use different features and datasets. In general, AI systems are designed and developed end-to-end in a siloed nature which prevents the reusing of features across projects and even sub-projects within the same project.
In this project, we are developing and validating a toolset, Data Extraction Suite, that provides a collaborative environment for data scientists, engineers, and clinical experts to define and manage reusable features and datasets from those features over FHIR-based data models for AI problems. These definitions include both rich metadata and scripts in FHIR search query and FHIR Path expression languages, when executed can extract the value of the feature. A data extraction engine is also provided as a part of the suite which can be configured and executed for fast and efficient dataset extraction that can be integrated with a range of machine learning environments in different ways e.g., writing to a Parquet file, or a feature stream or database (e.g., Feast, Amazon Sagemaker). With this decoupling, feature preparation becomes both FAIR, as users can search and access the metadata definition for features, and reproducible and reusable as anyone can connect its own data infrastructure via the FHIR API and extract the features ready for AI platforms to use automatically by executing the dataset definitions.
Tuncay Namlı, PhD. & Ali Anıl Sınacı, PhD.
Senior Researchers, Software Research & Development Company Corp.
Similar Posts
 Attitudes Towards the Adoption of Remote Patient Monitoring and Artificial...
Attitudes Towards the Adoption of Remote Patient Monitoring and Artificial...
 AI developments in healthcare. From models to application
AI developments in healthcare. From models to application

